Figma for Science
There might be a big startup opportunity with porting old, widely used scientific tools to the web with Figma-like realtime annotation and rendering capabilities.
Scientific tooling is old but collaboration's more important than ever.
Realtime thinking is important. Beautiful rendering is important.
There might be a common toolkit that builds figma-like tools and can be turned bespoke for particular domains. Widgets for different kinds of data displays and manipulation. Jupyter/Julia notebook style.
I’m especially interested in cell stains/histology/pathology because microscopy is gorgeous to look at, and seems underexplored.
https://axial.substack.com/p/axial-benchling
Existing work:
(Benchling)
- Benchling
- HASH
- FloydHub
- Streamlit
- NextJournal
- Biodock
- ScienceIO
- https://blockpad.net/
- https://getluminal.com/
- https://www.colabra.app/
- https://latchbio.substack.com/p/crispr-bioinformatics-in-the-browser
- https://latch.bio
- BenchSci
- https://kanobo.com/
- https://www.microns-explorer.org/cortical-mm3
- https://riboviz.org
- https://www.paige.ai
- https://www.openmicroscopy.org/omero/
- https://synbiohub.org
- https://www.radix.bio/
- https://simularium.allencell.org
- https://czid.org
- http://vis.nucleome.org/entry/examples
- https://www.jkanche.com/kana/
- https://pathpresenter.net/login
- https://pathcore.com/live
- https://twitter.com/michaellee04/status/1557738197068656640?s=21&t=ymJUaWO2ZBrj4HCj9oFDxQ
- https://www.byterat.io/
(Biodock)
"Experiment tracking" is a common theme
What if open science framework caught up with the times?
Journal of Open Research Software
An instance of Figma for X. Or maybe Palantir for X.
Local-only systems (figma-izable)
Geology/Space
Neuro
- https://github.com/MouseLand/receptivefield-explorer
- https://twitter.com/tomasfiers/status/1010580586208153600?s=21
- https://mne.tools/stable/index.html
- SimBrain
- https://surfer.nmr.mgh.harvard.edu/
- FSL
- https://www.janelia.org/lab/stringer-lab
Imagery
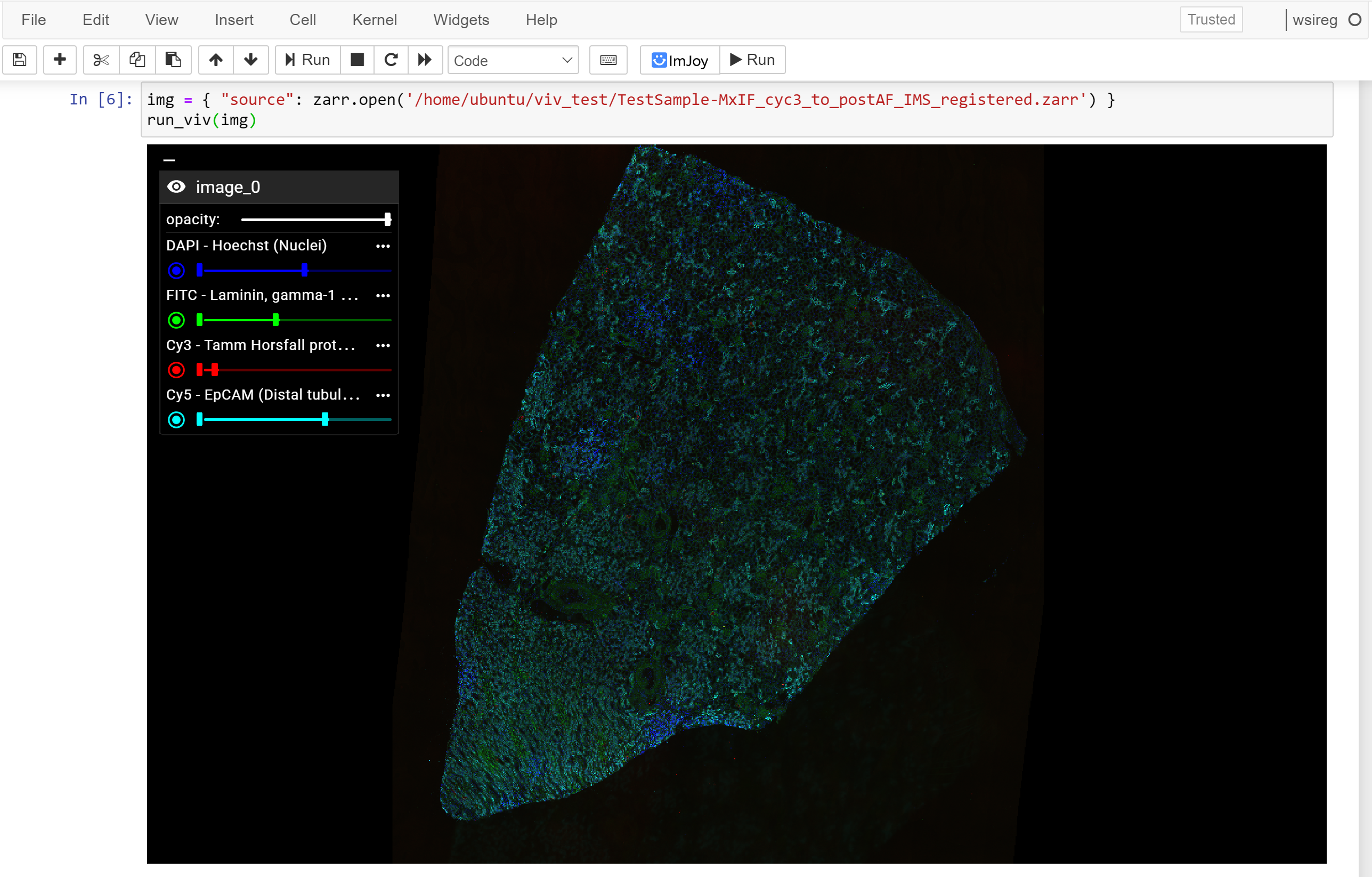
(Vizarr)
-
Bioformats - interop https://docs.openmicroscopy.org/bio-formats/6.8.1/about/index.html
-
ImageTwin (duplicate recognition) https://imagetwin.ai
Flow Cytometry
- https://www.flowjo.com/solutions/flowjo ($2000, on a dongle!)
- https://floreada.io/flow-cytometry-software
- https://www.beckman.com/flow-cytometry/software
Histology
The problem with microscopy is that file sizes are huge. Pathology/Histology is more tractable.
Informatics
Huge list here https://blog.addgene.org/free-online-molecular-biology-tools
Abstract Viz
Genomes
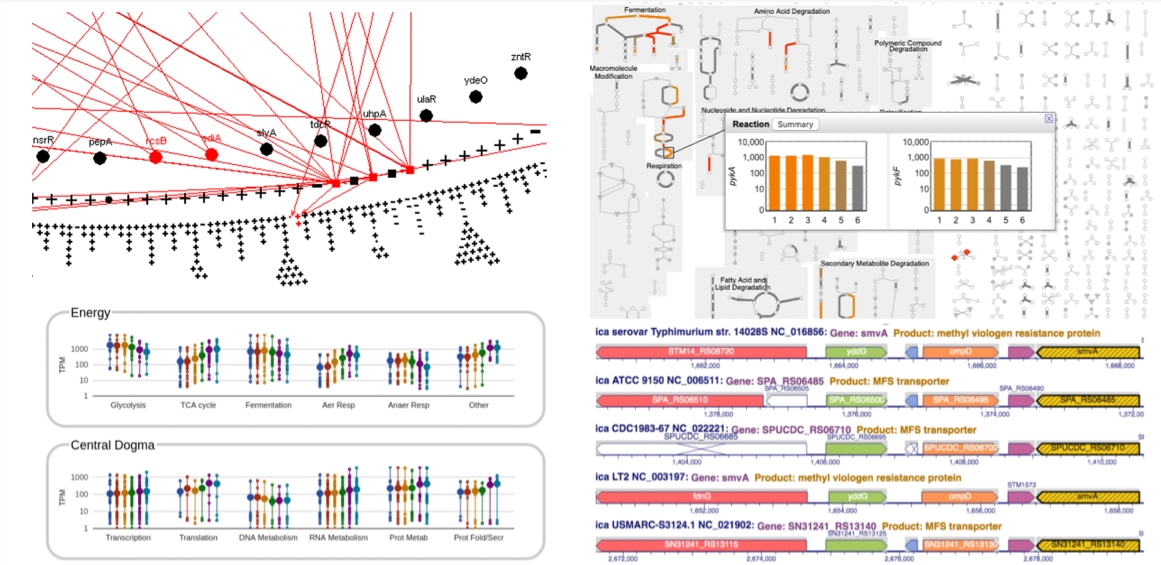
(Biocyc)
- https://biocyc.org
- https://jbrowse.org/jb2/
- http://gmod.org/wiki/GBrowse
- https://genomearchitect.readthedocs.io/en/latest/
- https://paintshop.io
- R plugin
- https://github.com/cov-lineages/pangolin
- https://biowasm.com
- https://aidenlab.org/juicebox/
- http://www.celavi.pro
- https://pangenome.github.io/
- https://higlass.io/about
Primers
Synthetics
- https://async.ece.utah.edu/tools/ibiosim/
- http://synbiocad.org
- http://cellocad.org/#/welcome?redirect=%2Fhome
Plasmid Editing
Great overview: https://medium.com/@alice.menglh/may-7th-13th-8ab68cbc9e7b
- https://www.snapgene.com
- http://serialbasics.free.fr/Serial_Cloner.html
- https://jorgensen.biology.utah.edu/wayned/ape/
- Open Vector Editor (https://github.com/TeselaGen/ove-electron)
- SeqViz (just a viewer) https://github.com/Lattice-Automation/seqviz
- https://www.geneious.com
- https://www.nucleobytes.com/enzymex/index.html
Protein/Mol Sim
Molecular Viz
- Gelbox
- http://cadnano.org
- http://crystalmaker.com
- CellPAINT
- https://pymol.org/2/
- http://www.yasara.org
- https://www.cgl.ucsf.edu/chimera/download.html
- "Blender for Biochemists"
- https://aip.scitation.org/doi/full/10.1063/1.5039473?Track=APLBJUN20
- http://iqmol.org
- https://clarafi.com/training/intro-mmaya-dsdna-kit/
Sim
Ligand Docking
In general, many domain specific tools are MATLAB plugins.
Molecule design
Misc
-
RCSB's PDB "view"s are crucial
-
http://blog.booleanbiotech.com/computational-tools-2022.html
DSP Notebook
Graphics
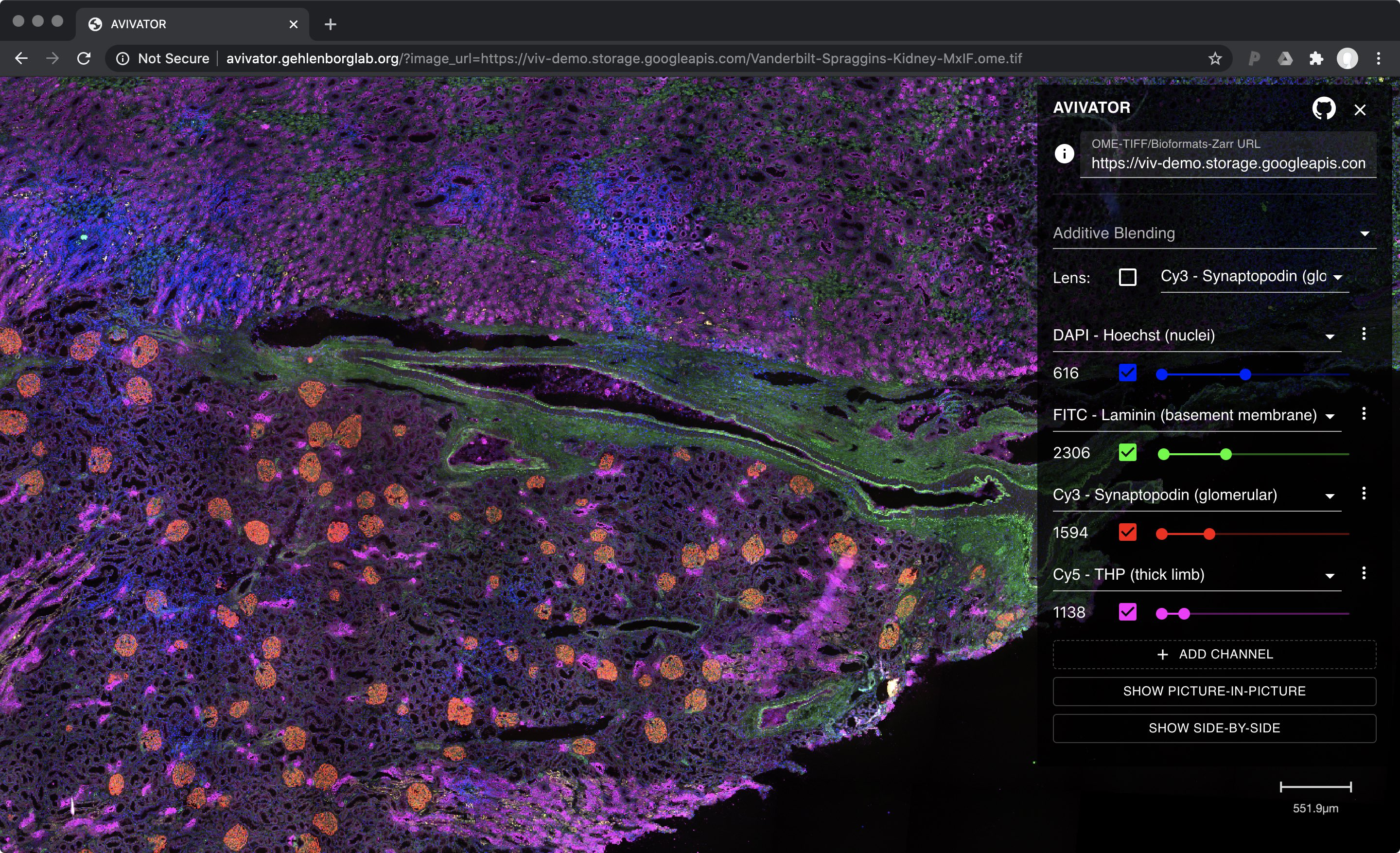
Because all rendering of high-bit depth #bioimage data is done via #WebGL shaders directly in the browser, Viv applies data transformations without lag. Magic lenses that perform local transformations are smooth and changes to the color mapping update as sliders are moved. pic.twitter.com/7AFD5cWObX
— Nils Gehlenborg (@ngehlenborg) August 13, 2020
Open source ethics
Data should be easy to sync to a repo in a common format
Really what's being provisioned is the collaboration tool and hosting. Reasonable to charge a price that would otherwise require on prem and manual setup (and social features/external sharing!)
—-
https://newscience.org/how-software-in-the-life-sciences-actually-works-and-doesnt-work/
Children
Backlinks